Research Article
Comparison and Improvement of Total RNA Extraction Methods from Different Tissues of Polygonatum cyrtonema Hua. 
2 College of Binjiang, Zhejiang Chinese Medical University, Hangzhou, 310053, China
3 Taizhou Traditional Chinese Medicine Hospital, Taizhou, 318020, China
 Author
Author  Correspondence author
Correspondence author
Medicinal Plant Research, 2023, Vol. 13, No. 1 doi: 10.5376/mpr.2023.13.0001
Received: 17 Apr., 2023 Accepted: 26 Apr., 2023 Published: 10 May, 2023
Zhang Y., Luo X.M., Luo X.J., Zhang S.L., Wang H., Zhang C.C., and Fan H.Y., 2023, Comparison and improvement of total rna extraction methods from different tissues of Polygonatum cyrtonema Hua., Medicinal Plant Research, 13(1): 1-9 (doi: 10.5376/mpr.2023.13.0001)
Extraction and isolation of high-quality RNA from Polygonatum cyrtonema Hua. is the basis of research on gene expression, regulation and genetic engineering. For screening the best method of total RNA extraction from different tissues of P. cyrtonema, total RNA was extracted from rhizomes, stems, leaves and flowers of P. cyrtonema by six methods that were Trizol method, CTAB-isopropanol method, RNA pure plant kit CTAB-LiCl method, hot phenol method and improved hot phenol method respectively. The concentration and quality of RNA were analyzed using Subordinate function method. The results showed that the improved hot phenol method was the most ideal for RNA extraction among the six methods. The RNA bands of different tissues of P. cyrtonema were complete and clear, the OD260/OD280 and OD260/OD230 values were between 1.8 and 2.1, the extraction concentration values were between 77.16 and 185.72 µg/g. This study provide a reference for extracting high-quality total RNA from P. cyrtonema.
Polygonatum cyrtonema Hua. is a perennial herb of the genus Polygonatum in the family of Liliaceae, which has ornamental, edible, medicinal and cosmetic values (Zhou et al., 2013), and is one of the unique medicinal plants with great development prospects in China. P. cyrtonema is widely used in folk medicine, with functions such as tonifying qi and nourishing yin, invigorating spleen, moistening lung, and tonifying kidney. It is known as the "King of Blood and Qi Tonifying" (Peng et al., 2017) and can be used clinically for diseases such as deficiency of spleen qi and stomach qi, tiredness and fatigue, stomach-yin deficiency, dry mouth without appetite, lung deficiency and dry cough, and soreness and weakness of waist and knees (Wang et al., 2015; Chen et al., 2015; Luo et al., 2016). With the widespread research on the medicinal value of P. cyrtonema, its artificial cultivation research has received widespread attention. The analysis of gene expression and gene transcriptome of key enzymes in the biosynthesis pathway of secondary metabolites such as polysaccharide and flavonoids in P. cyrtonema can provide basis for the optimization of artificial planting conditions (Ke, 2013; Li et al., 2017). Molecular biology experiments require a large amount of high-quality RNA, but due to the high content of secondary metabolites such as polysaccharides in P. cyrtonema samples, commonly used RNA extraction methods cannot be effectively applied to extract total RNA from different tissues of P. cyrtonema. And the commercial kits are not only expensive, but also have poor quality of extracted total RNA.
Subordinate function is a fundamental concept in fuzzy mathematics, which is widely used for comprehensive evaluation of crop stress resistance such as drought, salt alkali, shade, and waterlogging resistance (Liu et al., 2010). The evaluation of RNA quality is generally based on the analysis of indicators such as OD260/OD280, OD260/OD230, and RNA yield, lacking comprehensive evaluation methods. Due to the gradual variation and fuzziness of various evaluation indicators for RNA quality, applying the subordinate function method in fuzzy mathematics to evaluate RNA quality can obtain scientific evaluation results. In this study, the rhizomes, stems, leaves, and flowers of P. cyrtonema were used as materials to extract total RNA by six methods that were Trizol method, CTAB isopropanol method, Tiangen RNAprep Pure plant kit, hot phenol method, CTAB LiCl method, and improved hot phenol method. Subordinate function method was used to assist in comparing the integrity and yield of total RNA obtained by different extraction methods, and an economical, fast, and high-quality extraction method for total RNA from various tissues of P. cyrtonema was preferred to provide a reference for the subsequent molecular biology experiments of P. cyrtonema.
1 Results and Analysis
1.1 Quality analysis of total RNA extracted from P. cyrtonema using different methods
In this study, the integrity, degradation and pollution of total RNA extracted by different methods can be analyzed by observing the bands after agarose gel electrophoresis. The agarose gel electrophoresis patterns of RNA extracted from different tissues of P. cyrtonemawith different methods were obtained (Figure 1). The RNA bands extracted by CTAB-isopropanol method are blurry, with poor integrity, and DNA contamination, indicating significant degradation. The RNA bands of the leaves and stems of P. cyrtonema extracted by CTAB-LiCl method are clear, and the flower RNA bands have tail dragging phenomenon. At the same time, there is DNA contamination in each band. The electrophoretic bands of leaves, flowers, and stems using the hot phenol method are relatively bright and clear, indicating good integrity, but with slight tailing and DNA contamination. The RNA bands extracted from leaves and flowers using the Tiangen RNAprep pure plant kit method showed obvious trailing phenomena, indicating that they may have slight degradation. The RNA bands extracted by the Trizol method are blurry, indicating a low concentration and degradation. The improved hot phenol method extracted RNA from different parts of P. cyrtonema with clear and bright bands of 28S, 18S, and 5S, indicating that the RNA extracted by the improved hot phenol method had good integrity and no obvious degradation phenomenon.
|
Figure 1 Agarose gel electrophoresis showing total RNA extracted by 6 different methods Note: A: CTAB-sopropanol method; B: CTAB-LiCl method; C: Hot Phenol Method; D: Tiangen RNAprep pure plant kit; E: Trizol protocol; F: Improved hot phenol method; 1: Leaf; 2: Rhizome; 3: Flower; 4: Stem; a: 3-year-old rhizome; b: 2-year-old rhizome; c: 1-year-old rhizome |
1.2 Analysis of purity and yield of total RNA extracted from P. cyrtonema using different methods
The results of the purity and concentration data of RNA extracted from different tissues of P. cyrtonema using different methods (Table 1) showed that the purity of RNA extracted from different parts using CTAB-isopropanol method was not ideal, and the yield of RNA extracted from leaves, flowers, and stems was low. The CTAB-LiCl method had a good effect on RNA extraction from leaves and stems, with OD260/OD280 and OD260/OD230 values around 2.0. However, the yield of extracted RNA from roots, stems, and flowers was relatively low. The OD260/OD230 values of RNA extracted by the hot phenol method were both greater than 2.2, and the yield of extracted RNA from roots and stems was low. The purity and concentration of RNA extracted from leaves, rhizomes, and stems using the Trizol method were not ideal, with stem concentrations as low as 12.9 µg/g. The OD260/OD280 values of RNA extracted by the Tiangen RNAprep Pure plant kit method were between 1.8 and 2.1, but the yield of extracted RNA was low, with the yield of stem RNA as low as 19.6 µg/g. All the above methods were poorly extracted, while the improved hot phenol method extracted RNA with high purity and OD260/OD280 and OD260/OD230 values in the range of 1.8~2.1. Moreover, the yield of RNA was relatively high, with extraction concentrations ranging from 77.16 to 185.72 µg/g, which was suitable for extracting total RNA from different tissues of P. cyrtonema.
|
Table 1 Total RNA extracted from different tissues of Polygonatum cyrtonema Hua using different methods |
The results of ANOVA (Analysis of variance) showed that there were extremely significant differences in OD260/OD280, OD260/OD230, and RNA yield among different extraction methods (p<0.01). On OD260/OD280, there were significant differences between the CTAB-LiCl and the Tiangen RNAprep pure plant kit methods compared to the improved hot phenol method. Compared with the Trizol method, there were significant differences between the CTAB-LiCl and the Tiangen RNAprep pure plant kit method. On OD260/OD230, there were significant differences between the hot phenol method and the other four extraction methods except for the CTAB-LiCl method. The improved hot phenol method and the hot phenol method showed extremely significant differences compared to the Tiangen RNAprep pure plant kit method. It was found that there was a significant difference between the improved hot phenol method and the Tiangen RNAprep pure plant kit according to the RNA yield (Table 2).
|
Table 2 Total RNA extracted from different tissues of Polygonatum cyrtonema Hua using different methods Note: *: compared with CTAB-sopropanol method; #: compared with CTAB-LiCl method; $: compared with hot phenol method; △: compared with Trizol protocol; ※: compared with Tiangen RNAprep pure plant kit; ▲: compared with improved hot phenol method; Single symbol shows significant difference, p<0.05; Two repeating symbols show highly significant difference, p<0.01 |
1.3 Comprehensive analysis of total RNA extracted by different methods using subordinate function method
Subordinate function values of six extraction methods were obtained by calculating (Table 3). The larger the average value of the subordinate function, the higher the quality of RNA extracted under this extraction method. The analysis results showed that the order of quality of the extraction methods on OD260/OD280 was as follows: CTAB-LiCl method>improved hot phenol method>hot phenol method>Tiangen RNAprep pure plant kit>Trizol method>CTAB-isopropanol method. The extraction methods order on OD260/OD230 was as follows: improved hot phenol method>hot phenol method>CTAB-LiCl method>CTAB-isopropanol method>Trizol method>Tiangen RNAprep pure plant kit. And the extraction methods order on RNA yield was as follows: hot phenol method>improved hot phenol method>CTAB-isopropanol method>Trizol method>CTAB-LiCl method>Tiangen RNAprep pure plant kit.
|
Table 3 The membership function value of different extraction methods |
Based on the evaluation results of OD260/OD280, OD260/OD280, and RNA yield, the average subordinate function value of the total RNA quality extracted by the improved hot phenol method was the highest (0.764); The average subordinate function value of the Tiangen RNAprep pure plant kit was the lowest (0.504). The order was as follows: improved hot phenol method>hot phenol method>CTAB-LiCl method>CTAB-isopropanol method>Trizol method>Tiangen RNAprep pure plant kit.
1.4 Comparative analysis of the cost of the total RNA extraction from P. cyrtonema using different methods
By comprehensively comparing the operational complexity, experimental duration, economic cost, and overall RNA integrity and yield of six RNA extraction methods, including CTAB method, Trizol method, and so on, the advantages and disadvantages of the six methods were analyzed (Table 4). The Trizol method, as a traditional RNA extraction process with simple operation and low time cost, can effectively extract RNA from the flowers and stems of P. cyrtonema. However, due to the abundant content of secondary metabolites in the leaves and rhizomes of P. cyrtonema, the total RNA extracted by this method has poor integrity and low yield. CTAB-LiCl method is one of the traditional processes for extracting plant tissue RNA, which has low economic costs but is trifling and time-consuming to operate. Probably due to the multiple and time-consuming steps, RNA extraction was prone to degradation, and the effectiveness of this method in extracting leaves, rhizomes, and flower parts of P. cyrtonema was not satisfactory. Compared with the CTAB-LiCl method, the CTAB-isopropanol method has different RNA precipitation steps. Using isopropanol to precipitate RNA reduces the time and cost of extraction, but the extraction effect is still poor. This method extracts RNA from different parts of P. cyrtonema with poor integrity and low yield. The hot phenol method is often used to extract plant tissue RNA, which has low economic cost and simple operation steps, but the time cost is relatively high. And due to the rich content of polysaccharide and polyphenol products in the rhizome of P. cyrtonema, the RNA yield in the rhizome of P. cyrtonema extracted by this method is relatively low. Tiangen RNAprep pure plant kit is suitable for extracting RNA from plant tissues rich in secondary metabolites. Its operation is moderately cumbersome and takes a short time, but its economic cost is high. In practical applications, the effect of RNA extraction from different parts of P. cyrtonema is average and the yield of RNA from roots and stems is low. The improved hot phenol method adds further purification steps for RNA based on the hot phenol method. The operation is generally cumbersome, and the economic cost is low. The RNA extracted from different tissues of P. cyrtonema by this method has good integrity and high purity and yield.
|
Table 4 Comparison of different RNA extraction methods |
2 Discussion
Extracting high-quality RNA is a key step in analyzing gene expression, and an indispensable step in studying transcriptome sequencing, RT-PCR and other subsequent molecular research (Tong et al, 2012; Kim et al, 2014). P. cyrtonema is a typical plant material rich in secondary metabolites of polysaccharides and polyphenols, especially in its rhizome. It is difficult to extract high-quality RNA from this type of plant (Wang et al., 2005), and the content of secondary metabolites in different tissue parts varies (Ainsworth, 1994), further affecting the extraction efficiency of high-quality RNA from different parts of P. cyrtonema. As a rhizome medicine with a large demand but a small scale of artificial planting, the gene expression and transcriptome analysis of key enzymes in the biosynthesis pathway of polysaccharide, flavonoids and other secondary metabolites of P. cyrtonema can provide a basis for the optimization of artificial planting conditions. However, research on the molecular biology of the original plants of traditional Chinese medicinal materials such as P. cyrtonema is still in initial stage, and there is no research on RNA extraction methods for different tissues of P. cyrtonema. The main problem in the process of RNA extraction is degradation and contamination. In order to obtain RNA with good integrity from P. cyrtonema tissue rich in polysaccharides and polyphenols, the most crucial thing is that the fragmentation of plant cells must be complete, sufficient, and rapid (Wei, 2019).
In this study, six methods of Trizol method, CTAB-isopropanol method, CTAB-LiCl method, hot phenol method, Tiangen RNAprep pure plant kit and improved hot phenol method were used to extract total RNA from the rhizome, stem, leaf, and flower parts of P. cyrtonema, and analyzed using subordinate function method. The results showed that the quality of RNA extracted by the Trizol method was poor, possibly due to its irreversible binding with polyphenols and nucleic acid macromolecules, resulting in RNA loss. The yield of RNA extracted from leaves, flowers and stems by CTAB-isopropanol method was low and the integrity was extremely poor, which may be caused by the precipitation of RNA by isopropanol, which led to the coprecipitation of polysaccharides and phenols. The CTAB-LiCl method was prone to RNA degradation due to its overly trivializing operational steps and time-consuming time costs, resulting in low yield and poor quality of RNA extracted in practical operations. The economic cost of the kit method was the highest, and the excessive content of polysaccharides in P. cyrtonema resulted in low RNA extraction yield and poor integrity. Although the concentration of P. cyrtonema leaves, flowers, and stem tissues extracted by the hot phenol method was high, there was DNA contamination and the integrity of RNA extracted from the root and stem tissues was poor. The RNA extracted by the improved hot phenol method not only had good integrity, but also had high quality, which met the conditions for extracting RNA from different parts of P. cyrtonema. It could be well used for subsequent related molecular biology experiments. The improved hot phenol method modified the RNA precipitant and added purification steps based on the hot phenol method, using isopropanol to precipitate RNA, saving time and cost. Subsequently, RNA was re-precipitated with 3 mol/L pH5.2 sodium acetate and anhydrous ethanol. The increased purification steps reduced the side effects of coprecipitation of polysaccharides and phenols caused by isopropanol precipitation of RNA and improved the quality of RNA. The application of subordinate function method could comprehensively evaluate the quality of total RNA extracted by different methods. The larger the average subordinate function value, the better the quality of total RNA extracted. Conversely, the worse the quality. The analysis results indicated that the quality of total RNA extracted by different methods was ranked as follows: improved hot phenol method>hot phenol method>CTAB-LiCl method>CTAB-isopropanol method>Trizol method>Tiangen RNAprep pure plant kit. In a word, the improved hot phenol method was suitable for extracting high-quality RNA from different tissues of P. cyrtonema.
3 Materials and Methods
3.1 Samples
The young leaves, tender stems, rhizomes, and flowers of the tested P. cyrtonema were all purchased from the Planting Base of Yikang Agriculture and Forestry Technology Co., Ltd. in Qingyuan County. It was identified by Professor Zhang Shuili from the College of Pharmaceutical Sciences, Zhejiang Chinese Medical University as a plant of Polygonatum cyrtonema Hua, belongs to Polygonum genus in the family of Liliaceae. After being frozen in liquid nitrogen, the sample was stored in a refrigerator at -80 °C for future use.
3.2 Instruments and reagents
The HH420 constant temperature water bath was purchased from Shangyu Daoxu Maoxiang Instrument Equipment Factory. MiniT-H2C triple multi-purpose metal bath was purchased from Hangzhou Allsheng Instruments Co., Ltd. High-speed freezing centrifuge (Beckman 64R Centrifuge) was purchased from Beckman Coulter, Inc. (USA). Vortex mixers XW-80A was purchased from Haimen Kylin-Bell Lab Instruments Co., Ltd. Nanodrop 2000 Ultra-micro spectrophotometer was purchased from Thermo Fisher Scientific. DYY-8C double stable electrophoresis instrument was purchased from Beijing Liuyi Instrument Factory. Master cycler Pro S-Gradient PCR instrument was purchased from Eppendorf (Germany). Tanon 4100 automatic digital gel imaging analysis system was purchased from Shanghai Tanon Technology Co., Ltd.
Trizol reagent and isoamyl alcohol were purchased from Ambion and Shanghai Chemical Reagent Co., Ltd. Reagent Factory 1, respectively. Tris-HCl, Sodium dodecyl sulfate (SDS) and Hexadecyl trimethyl ammonium Bromide were purchased from Amresco (USA). Ethylene Diamine Tetraacetic Acid (EDTA), 2-Hydroxy-1-ethanethiol, polyvinyl pyrrolidone (PVP), Spermidine, Tris(trimethylsiloxy)boron, Trichloromethane, buffer solution, Tiangen RNAprep pure plant kit, Sodium acetate, Diethyl pyrocarbonate (DEPC), and Ethanol absolute were purchased from Shanghai Chemical Reagent Purchase Supply Wulian Chemical Plant, Beijing Dingguochangsheng BIOTECHNOLOGY Co., Ltd., Sigma-Aldrich (USA), Nanjing Aoduo Funi Biology Technology Co., Ltd., Tiangen Biotech Co., Ltd., Xilong Scientific Co., Ltd., Beijing Yingcheng Jiye Chemical Co., Ltd., Tiangen Biotech Co., Ltd., Wenzhou Runhua Chemical Industry Co., Ltd., Beijing Solarbio Science & Technology Co., Ltd., Hangzhou Longshan Fine Chemical Co., Ltd, respectively. Lithium chloride and propan-2-ol (IPA) were purchased from Tianjin Kermel Chemical Reagent Co., Ltd.
3.3 Experimental methods
Trizol method: Refer to the method of Wu et al. (2012). (1) Take 200~300 mg of P. cyrtonema tissue and grind it into powder in liquid nitrogen. Quickly add 600 µL Trizol reagent, mix well, and place it in an ice bath for 5 min. Centrifuge at 4 °C and 12 000 r/min for 10 min. Then, take the supernatant into a new centrifuge tube, add one-fifth volume of trichloromethane, mix well, and then ice bath for 5 minutes. Centrifuge for 20 min at 4 °C and 12 000 r/min. (2) Take the supernatant and place it in a new centrifuge tube. Add an equal volume of pre-cooled isopropyl alcohol, ice bath for 10 min at 4 °C and 12 000 r/min, and centrifuge for 10 min. Remove the supernatant and leave sediment. (3) Add 8.0 mol/L LiCl solution to the supernatant to achieve a final concentration of 2.0 mol/L, and place at 4 °C overnight. Then, centrifuge at 4 °C and 12 000 r/min for 10 minutes. Remove the supernatant, wash the precipitate with 75% ethanol, add 500 µL of SSTE buffer to dissolve the precipitate, and then add 500 µL of chloroform: isoamyl alcohol (24:1) and extract to completely precipitate anhydrous ethanol. (4) Wash the precipitate with 1 mL of 75% ethanol (prepared with DEPC and pre-cooled at 4 °C) and centrifuge at 4 °C and 12 000 r/min for 3 min. Remove the waste liquid, dry and dissolve the precipitate in 40 µLDEPC. Final, store it at -80 °C for future use.
CTAB-LiCl method: Refer to the method of Meng et al. (2016). (1) Take 200~300 mg of P. cyrtonema tissue and grind it into powder in liquid nitrogen. Quickly add 600 µL 65 °C preheated of CTAB extraction solution, and immediately vortex oscillate. Take a 65 °C water bath for 5 min and oscillate 3~5 times during this period. (2) After cooling, add equal volume of chloroform/isoamyl alcohol and shake well. Centrifuge at room temperature at 12 000 r/min for 15 min, then transfer the supernatant to a new centrifuge tube. (3) Repeat the operation of (2). (4) Add (1/3 volume) 8 mol/L LiCl to the supernatant transferred to the new centrifuge tube, resulting in a final concentration of 2 mol/L, and overnight at 4 °C. (5) Centrifuge at 4 °C and 10 000 r/min for 20 min and remove the supernatant. Wash the precipitate with 1 mL of 70% ethanol, air dry, add 500 µL of SSTE buffer to dissolve the precipitate, then add 500 µL of chloroform/isoamyl alcohol again, and extract twice. Add 1 mL of anhydrous ethanol to the supernatant and place it at -70 °C for more than 30 min. (6) Centrifuge at 4 °C and 12 000 r/min for 20 min. Wash the precipitate with 1 mL of 70% ethanol and 1 mL of anhydrous ethanol, dry and dissolve the precipitate with 40 µL of DEPC.
CTAB-isopropanol method: Refer to the method of "CTAB-LiCl method", and the RNA precipitation step is changed to "adding equal volume of isopropanol, mixing well, and standing on ice for 30 min".
Hot phenol method: Refer to the method of Si et al. (2012). (1) Take 200~300 mg of P. cyrtonema tissue, grind with liquid nitrogen, add 1 mL of 65 °C preheated extraction solution, vortex shake and mix well. (2) Add 1/2 volume of chloroform/isoamyl alcohol (24:1 solution), shake and mix well, and centrifuge for 15 min at 4 °C and 12 000 r/min. (3) Take the supernatant and place it in a new centrifuge tube. Add 1/3 volume of 10 mol/L LiCl to the supernatant and place at -20 °C for 1.5 h. (4) Centrifuge at 4 °C and 12 000 r/min for 30 min and remove the supernatant. Suspend the precipitate with a concentration of 500 µL of 200 mmol/L NaCl, add 500 µLof buffer solution, shake well, and ice bath for 10 min. (5) Centrifuge at 4 °C and 12 000 r/min for 15 min, then take the supernatant and place it in a new centrifuge tube. Add an equal volume of chloroform/isoamyl alcohol (24:1) solution, mix well, and then take an ice bath for 10 min. (6) Centrifuge at 4 °C and 12 000 r/min for 10 min. Take the supernatant into a new centrifuge tube, add 2 times volume of anhydrous ethanol, and place at -20 °C for 30 min. (7) Centrifuge at 4 °C and 12 000 r/min for 10 min. Remove the supernatant, wash the precipitate twice with 75% ethanol, dry on clean bench, dissolve in 40 µL DEPC water, and store at -80 °C for later use.
Tiangen RNAprep pure plant kit method: Refer to the instructions of this kit.
Improved hot phenol method: (1) Preheat a mixed solution of 400 µL phenol water and 400 µL RNA extraction buffer (0.1 mol/L Tris-HCl (pH 8.0), 0.1 mol/L LiCl, 0.01 mol/L EDTA (pH 8.0), 1.0% SDS) in an 80 °C metal bath. (2) Thoroughly grind 200~300 mg of P. cyrtonema sample in liquid nitrogen, and transfer the sample to a 1.5 mL pre-cooled centrifuge tube, then quickly add a preheated 80 °Cmixed solution, mix vigorously, add 400 mL of chloroform, and place on the ice for 15 min. (3) Centrifuge at 4 °C and 12 000 r/min for 20 min, take the supernatant and place it in a new centrifuge tube. Add equal volume of isopropanol and mix well. Place it on ice for 30 min. (4) Centrifuge at 4 °C and 12 000 r/min for 20 min, remove the supernatant. Add 360 mL DEPC, 40 mL of 3 mol/L sodium acetate (pH 5.2), and 800 mL of anhydrous ethanol, mix well, and place at -20 °C for 2 h. (5) Centrifuge at 4 °C and 12 000 r/min for 15 min, remove the supernatant. (6) RNA precipitation is washed once with 1 mL of 70% and 100% ethanol, respectively. After drying on clean bench, it is dissolved in 40 mL DEPC and stored in a -80 °C refrigerator for future use.
Determination of total RNA purity and yield: Extracted 2 µL RNA samples and measured the RNA concentration, OD260/OD280, and OD260/OD230 values with the help of Nanodrop 2000 ultra micro spectrophotometer.
Analysis of total RNA integrity: The integrity of the extracted RNA was detected by 1.2% agarose gel electrophoresis. Precision suction 5 μL P. cyrtonema RNA sample, with 1 μL 10×Loading Buffer mix well and sample. The electrophoresis conditions were set as 120 V and 30 min. Electrophoresis results were photographed and recorded in the gel imaging system.
Data analysis: Statistical analysis of the data was conducted using Excel and SPSS 20.0 software, and the subordinate function method (Meng et al., 2006; Liu et al., 2010; Wu et al., 2012; Si et al., 2012; Xu et al., 2018) was used to comprehensively evaluate the total RNA yield and purity extracted by different methods. The evaluation index value showed an "S" curve relationship (positive correlation) with RNA yield, and a parabolic relationship with OD260/OD280 and OD260/OD230 values. The formula for calculating RNA yield (1) and the formula for calculating RNA OD260/OD280 and OD260/OD230 values (2) are as follows:
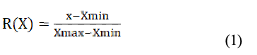
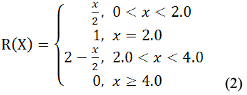
Where, x represents the measured value of the indicator, Xmax and Xmin represent the maximum and minimum RNA yield of all samples, respectively.
Authors’ contributions
ZY and LXM are the executors of the experimental operation, data analysis, and paper draft writing of this study. LXJ participated in experimental sampling and operation. ZSL, WH, and ZCC participated in experimental guidance and data analysis. FHY is the conceptualizer and leader of this project, guiding experimental design, data analysis, paper writing, and revision. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the Zhejiang Basic Public Welfare Research Project (LGN20C140003), Open Research Fund for Traditional Chinese Medicine Discipline of Zhejiang Chinese Medical University (Yao2016010), Science Research Innovation Fund for Young and Middle-aged Scholars of Zhejiang Chinese Medical University (KC201908) and Project of Zhejiang Traditional Chinese Materia Medica Resource Survey (17-21209). Thanks for the technical assistance provided by the Public Platform of the Medical Research Center of the School of Traditional Chinese Medicine, Zhejiang Chinese Medical University.
Ainsworth C., 1994, Isolation of RNA from floral tissue of Rumex acetosa (Sorrel), Plant Molecular Biology Reporter, 12(3): 198-203
https://doi.org/10.1007/BF02668741
Chen H., Feng S.S., Sun Y.J., Hao Z.Y., Feng W.S., and Zheng X.K., 2015, Advances in studies on chemical constituents of three medicinal plants from Polygonatum Mill. and their pharmacological activities, Zhongcaoyao (Chinese Traditional and Herbal Drugs), 46(15): 2329-2338
Ke H.M., 2013, The fate of key genes related to flavonoids biosynthetic way in resynthesized Brassicanapus, Thesis for M.S., Huazhong Agricultural University, Supervisor: Fu T.D., and Wen J., pp.18-32
Kim J.H., Jin H.O., Park J.A., Chang Y.H., Hong Y.J., and Lee J.K., 2014, Comparison of three different kits, for extraction of high-quality RNA from frozen blood, SpringerPlus, 3(1): 76
https://doi.org/10.1186/2193-1801-3-76
PMid:24567882 PMCid:PMC3925490
Li R.Q., 2017, Overexpression of key genes in biosynthesis pathway of ganoderma lucidum polysaccharide, Thesis for M.S., Jiangnan University Supervisor: Ding C.Y., pp.22-23
Liu X.Y., Liu X.Y., and Wang M., 2010, The determination of membership function and use, Diannao Zhishi Yu Jishu (Computer Knowledge and Technology), 6(31): 8831-8832
Luo M., Zhang W.W., Deng C.F., Tan Q.S., Luo C., and Luo S., 2016, Advances in studies of medicinal crop Polygonatum cyrtonema Hua, Lishizhe Guoyi Guoyao (Lishizhen Medicine and Materia Medica Research), 27(6): 1467-1469
Meng L., Zhou L., Zhang M.S., and Dai S.L., 2006, An efficient and economic method for preparation total rna of petals, Shengwu Jishu (Biotechnology), 16(1): 38-40
Peng X.B., 2017, Ecological and economic benefits of the combined management of chestnut and Polygonatum, Thesis for M.S., Journal of Central South University of Forestry and Technology, Supervisor: Yuan D. pp.1-2
Si A.J., Ruan M.B., and Xie Z.M., 2012, Rapid extraction of total RNA from different cotton tissues by modified hot-phenol method, Anhui Nongye Kexue (Journal of Anhui AgriculturalSciences), 40(16): 8830-8832
Tong Z.G., Qu S.C., Zhang J.Y., Wang F., Tao J.M., Gao Z.H., and Zhang Z., 2012, A modified protocol for RNA extraction from different peach tissues suitable for gene isolation and Real-Time PCR analysis, Mol. Biotechnology, 50(3): 229-236
https://doi.org/10.1007/s12033-011-9433-3
PMid:21744035
Wang T., and Miao M.S., 2015, The modern research and analysis of Polygonatum, Zhongyi Xuebao (Chinese Medical Journal), 30(5): 714-718
Wang T., Zhang N.H., and Du L.F., 2005, Isolation of RNA of high quality and yield from Ginkgo biloba leaves, Biotechnology Letters, 27(9): 629-633
https://doi.org/10.1007/s10529-005-3629-1
PMid:15977069
Wei H.Y., Wei W., Yang M., Zhao J.R., Zhong Y., Liu J.N., Chen Z.B., Pei W.H., Chen H.R, and Yu L., 2019, Comparative Studyon RNA Extraction Methods from Konjac (Amorphophal-lusspp.) of Four Organs, Fengzi Zhiwu Yuzhong (Molecular Plant Breeding), 17(21): 7064-7070
Wu K.C., Huang C.M., li Y.R., Yang L.T., and Wu J.M., 2012, Fast and effective total RNA extraction from different tissues in 3 crops through the Trizol reagent method, Nabfang Nongye Xuebao (Journal of Southern Agriculture), 43(12):1934-1939
Xu L., Zhao C.L., Zeng X.L., Zhang H.L., Pan Y.B., 2018, Subordinate Fu nction-assisted Comparison of Total RNA Extraction Methods of Aerial Stem from Annual Panax notoginseng, Fengzi Zhiwu Yuzhong (Molecular Plant Breeding), 16(1): 328-333
Zhou J.J., Luo X.F., Ye W., and Jiang J.L., 2013, Study on seed propagation techniques of Pcyrtonema Hua, Zhongzi (Seed), 32(1): 111-113
. PDF(713KB)
. HTML
Associated material
. Readers' comments
Other articles by authors
. Ying Zhang
. Xiaomeng Luo
. Xingju Luo
. Shuili Zhang
. Hong Wang
. Chunchun Zhang
. Huiyan Fan
Related articles
. Polygonatum cyrtonema Hua.
. RNA extraction
. Improved hot phenol method
. Subordinate function method
Tools
. Email to a friend
. Post a comment
.png)
.png)
.png)
.png)
.png)


